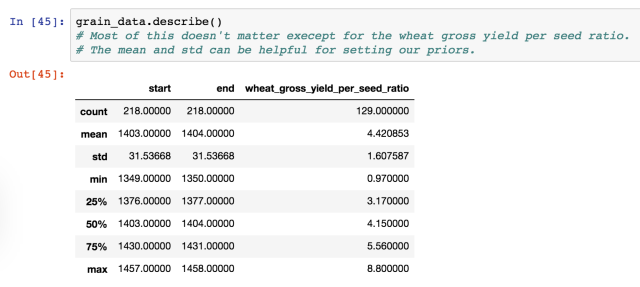
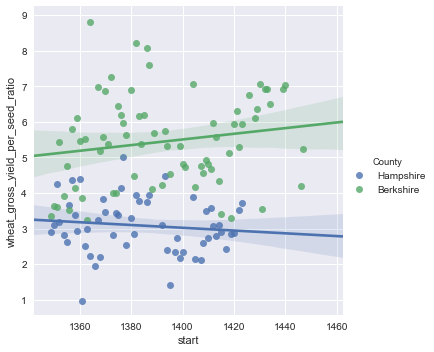
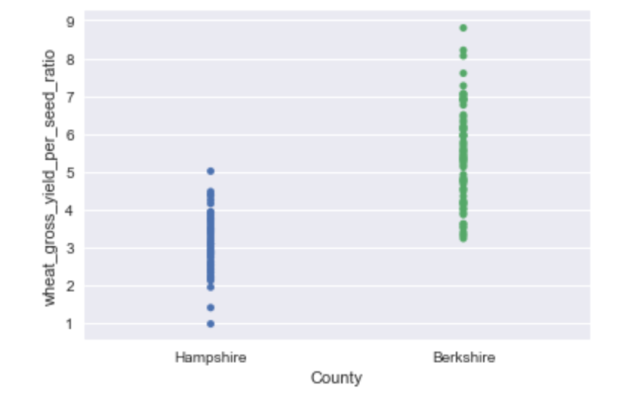
In the last post we talked about the data set up and writing the model for comparing crop yields from two different counties in England.
This will run the model for us.
# Compiling and producing posterior samples from the model. fit_compare_groups = pystan.stan(model_code=model_string, data=compare_groups, iter=1000, chains=4)
And now we can look at the model output.
fit_compare_groups
<pre>Inference for Stan model: anon_model_1f0ebaa1784e353e9595dea46e6ad140.
4 chains, each with iter=1000; warmup=500; thin=1;
post-warmup draws per chain=500, total post-warmup draws=2000.
mean se_mean sd 2.5% 25% 50% 75% 97.5% n_eff Rhat
mu[0] 5.44 3.4e-3 0.15 5.14 5.34 5.44 5.55 5.74 2000 1.0
mu[1] 3.08 2.5e-3 0.11 2.86 3.01 3.08 3.16 3.3 1930 1.0
sigma[0] 1.31 2.4e-3 0.11 1.11 1.23 1.31 1.38 1.54 2000 1.0
sigma[1] 0.8 1.8e-3 0.08 0.66 0.74 0.79 0.85 0.96 1974 1.0
lp__ -70.59 0.04 1.37 -74.0 -71.3 -70.3 -69.57 -68.82 1092 1.0
Samples were drawn using NUTS at Wed Jul 5 12:42:43 2017.
For each parameter, n_eff is a crude measure of effective sample size,
and Rhat is the potential scale reduction factor on split chains (at
convergence, Rhat=1).
Right off the we can that mu0 and mu1 are not the same. A few lines of code will help verify this.
# Plotting the posterior distribution fit_compare_groups.plot()
This is the trace plot from the model.
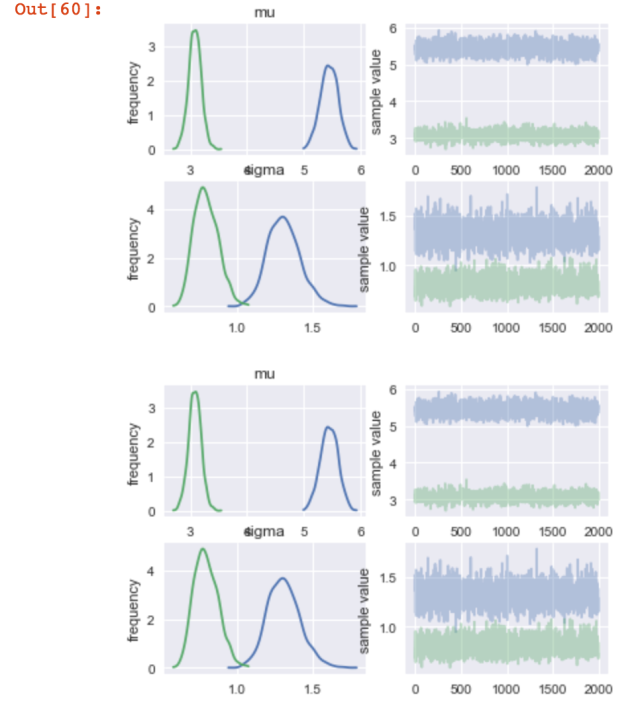
There are a few other things you can do as well.
# Returns the mean of the location paramter mu mu = fit_compare_groups.extract(permuted=True)['mu'] np.mean(mu, axis=0)
and
mu_df = pd.DataFrame(mu) mu_df.columns = ['mu0', 'mu1'] # The probability that mu0 is smaller than mu1 l = mu_df.mu0 < mu_df.mu1 sum(l)/len(l)